IV. Archive Classification¶
The primary question which we pursue in this section is how one can use reproducible and replicable workflows for discovering the optimal classifications of the text groups from the Drehem texts, found in an unprovenanced archival context. We describe how we leverage existing classification models to help validate our findings.
# import necessary libraries
import pandas as pd
from tqdm.auto import tqdm
# import libraries for this section
import re
import matplotlib.pyplot as plt
# import ML models from sklearn
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.decomposition import PCA
from sklearn.naive_bayes import BernoulliNB
from sklearn.naive_bayes import GaussianNB
from sklearn import svm
# import train_test_split function
from sklearn.model_selection import train_test_split
from sklearn import metrics
1 Set up Data¶
Create a dictionary of archive categories
1.1 Labeling the Training Data¶
We will be labeling the data according to what words show up in it.
labels = dict()
labels['domesticated_animal'] = ['ox', 'cow', 'sheep', 'goat', 'lamb', '~sheep', 'equid'] # account for plural
#split domesticated into large and small - sheep, goat, lamb, ~sheep would be small domesticated animals
labels['wild_animal'] = ['bear', 'gazelle', 'mountain'] # account for 'mountain animal' and plural
labels['dead_animal'] = ['die'] # find 'die' before finding domesticated or wild
labels['leather_object'] = ['boots', 'sandals']
labels['precious_object'] = ['copper', 'bronze', 'silver', 'gold']
labels['wool'] = ['wool', '~wool']
# labels['queens_archive'] = []
Using filtered_with_neighbors.csv generated above, make P Number and id_line the new indices.
Separate components of the lemma.
words_df = pd.read_pickle('https://gitlab.com/yashila.bordag/sumnet-data/-/raw/main/part_3_words_output.p') # uncomment to read from online file
#words_df = pd.read_pickle('output/part_3_output.p') #uncomment to read from local file
data = words_df.copy()
data.loc[:, 'pn'] = data.loc[:, 'id_text'].str[-6:].astype(int)
data = data.set_index(['pn', 'id_line']).sort_index()
extracted = data.loc[:, 'lemma'].str.extract(r'(\S+)\[(.*)\](\S+)')
data = pd.concat([data, extracted], axis=1)
data = data.fillna('') #.dropna() ????
data.head()
| lemma | id_text | id_word | label | date | dates_references | publication | collection | museum_no | ftype | metadata_source_x | prof? | role? | family? | number? | commodity? | neighbors | min_year | max_year | min_month | max_month | diri_month | min_day | max_day | questionable | other_meta_chars | multiple_dates | metadata_source_y | 0 | 1 | 2 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pn | id_line | |||||||||||||||||||||||||||||||
| 100041 | 3 | 6(diš)[]NU | P100041 | P100041.3.1 | o 1 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | Yes | No | [] | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | 6(diš) | NU | ||||
| 3 | udu[sheep]N | P100041 | P100041.3.2 | o 1 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | No | No | [] | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | udu | sheep | N | ||||
| 4 | kišib[seal]N | P100041 | P100041.4.1 | o 2 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | No | No | [] | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | kišib | seal | N | ||||
| 4 | Lusuen[0]PN | P100041 | P100041.4.2 | o 2 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | No | No | [6(diš)[]NU, udu[sheep]N, kišib[seal]N, Lusuen... | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | Lusuen | 0 | PN | ||||
| 5 | ki[place]N | P100041 | P100041.5.1 | o 3 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | No | No | [] | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | ki | place | N |
data['label'].value_counts()
o 1 46955
o 2 37850
o 3 36878
r 3 33824
o 4 32289
...
env o 10 1
o vii' 4 1
o ii 50 1
a i 9 1
seal S000081 ii 4 1
Name: label, Length: 2032, dtype: int64
for archive in labels.keys():
data.loc[data.loc[:, 1].str.contains('|'.join([re.escape(x) for x in labels[archive]])), 'archive'] = archive
data.loc[:, 'archive'] = data.loc[:, 'archive'].fillna('')
data.head()
| lemma | id_text | id_word | label | date | dates_references | publication | collection | museum_no | ftype | metadata_source_x | prof? | role? | family? | number? | commodity? | neighbors | min_year | max_year | min_month | max_month | diri_month | min_day | max_day | questionable | other_meta_chars | multiple_dates | metadata_source_y | 0 | 1 | 2 | archive | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pn | id_line | ||||||||||||||||||||||||||||||||
| 100041 | 3 | 6(diš)[]NU | P100041 | P100041.3.1 | o 1 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | Yes | No | [] | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | 6(diš) | NU | |||||
| 3 | udu[sheep]N | P100041 | P100041.3.2 | o 1 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | No | No | [] | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | udu | sheep | N | domesticated_animal | ||||
| 4 | kišib[seal]N | P100041 | P100041.4.1 | o 2 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | No | No | [] | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | kišib | seal | N | |||||
| 4 | Lusuen[0]PN | P100041 | P100041.4.2 | o 2 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | No | No | [6(diš)[]NU, udu[sheep]N, kišib[seal]N, Lusuen... | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | Lusuen | 0 | PN | |||||
| 5 | ki[place]N | P100041 | P100041.5.1 | o 3 | SSXX - 00 - 00 | SSXX - 00 - 00 | AAS 053 | Louvre Museum, Paris, France | AO 20313 | BDTNS | No | No | No | No | No | [] | 0 | 1 | False | 0 | 1 | False | False | False | BDTNS | ki | place | N |
The function get_set has a dataframe row as an input and returns a dictionary where each key is a word type like NU and PN. The values are its corresponding lemmas.
1.2 Data Structuring¶
def get_set(df):
d = {}
seals = df[df['label'].str.contains('seal')]
df = df[~df['label'].str.contains('seal')]
for x in df[2].unique():
d[x] = set(df.loc[df[2] == x, 0])
d['SEALS'] = {}
for x in seals[2].unique():
d['SEALS'][x] = set(seals.loc[seals[2] == x, 0])
return d
get_set(data.loc[100271])
{'': {''},
'MN': {'Šueša'},
'N': {'itud', 'maš', 'mu', 'mu.DU', 'udu'},
'NU': {'1(diš)', '2(diš)'},
'PN': {'Apilatum', 'Ku.ru.ub.er₃', 'Šulgisimti'},
'SEALS': {},
'SN': {'Šašrum'},
'V/i': {'hulu'},
'V/t': {'dab'}}
archives = pd.DataFrame(data.groupby('pn').apply(lambda x: set(x['archive'].unique()) - set(['']))).rename(columns={0: 'archive'})
archives.loc[:, 'set'] = data.reset_index().groupby('pn').apply(get_set)
archives.loc[:, 'archive'] = archives.loc[:, 'archive'].apply(lambda x: {'dead_animal'} if 'dead_animal' in x else x)
archives.head()
| archive | set | |
|---|---|---|
| pn | ||
| 100041 | {domesticated_animal} | {'NU': {'6(diš)'}, 'N': {'ki', 'kišib', 'udu'}... |
| 100189 | {dead_animal} | {'NU': {'1(diš)', '5(diš)-kam', '2(diš)'}, 'N'... |
| 100190 | {dead_animal} | {'NU': {'3(u)', '1(diš)', '5(diš)', '1(diš)-ka... |
| 100191 | {dead_animal} | {'NU': {'1(diš)', '4(diš)', '4(diš)-kam', '2(u... |
| 100211 | {dead_animal} | {'NU': {'1(diš)', '1(u)', '1(diš)-kam', '2(diš... |
def get_line(row, pos_lst=['N']):
words = {'pn' : [row.name]} #set p_number
for pos in pos_lst:
if pos in row['set']:
#add word entries for all words of the selected part of speech
words.update({word: [1] for word in row['set'][pos]})
return pd.DataFrame(words)
Each row represents a unique P-number, so the matrix indicates which word are present in each text.
sparse = words_df.groupby(by=['id_text', 'lemma']).count()
sparse = sparse['id_word'].unstack('lemma')
sparse = sparse.fillna(0)
sparse = pd.concat(archives.apply(get_line, axis=1).values).set_index('pn')
sparse
| ki | kišib | udu | itud | mu | ud | ga | sila | šu | mu.DU | maškim | ekišibak | zabardab | u | maš | šag | lugal | mašgal | kir | a | en | ensik | egia | igikar | ŋiri | ragaba | dubsar | mašda | saŋŋa | amar | mada | akiti | lu | ab | gud | ziga | uzud | ašgar | gukkal | šugid | ... | šaŋanla | pukutum | lagaztum | bangi | imdua | KU.du₃ | batiʾum | niŋna | sikiduʾa | gudumdum | šuhugari | šutur | gaguru | nindašura | ekaskalak | usaŋ | nammah | egizid | nisku | gara | saŋ.DUN₃ | muhaldimgal | šagiagal | šagiamah | kurunakgal | ugulaʾek | šidimgal | kalam | enkud | in | kiʾana | bahar | hurizum | lagab | ibadu | balla | šembulug | li | niŋsaha | ensi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pn | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 100041 | 1.0 | 1.0 | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 100189 | 1.0 | NaN | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 100190 | 1.0 | NaN | 1.0 | 1.0 | 1.0 | 1.0 | NaN | 1.0 | NaN | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 100191 | 1.0 | NaN | 1.0 | 1.0 | 1.0 | 1.0 | NaN | NaN | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | 1.0 | 1.0 | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 100211 | 1.0 | NaN | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1.0 | 1.0 | 1.0 | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 519650 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 519658 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 519792 | NaN | NaN | 1.0 | 1.0 | 1.0 | 1.0 | NaN | 1.0 | NaN | 1.0 | NaN | NaN | NaN | NaN | 1.0 | NaN | NaN | 1.0 | NaN | 1.0 | NaN | 1.0 | NaN | NaN | NaN | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1.0 | NaN | NaN | 1.0 | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 519957 | NaN | NaN | NaN | 1.0 | NaN | NaN | NaN | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 519959 | 1.0 | 1.0 | NaN | 1.0 | NaN | NaN | NaN | 1.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
15139 rows × 1076 columns
sparse = sparse.fillna(0)
sparse = sparse.join(archives.loc[:, 'archive'])
sparse.loc[sparse.loc[:, 'archive'].apply(lambda x: 'domesticated_animal' in x), 'domesticated_animal'] = 1
sparse.loc[:, 'domesticated_animal'] = sparse.loc[:, 'domesticated_animal'].fillna(0)
sparse.loc[sparse.loc[:, 'archive'].apply(lambda x: 'wild_animal' in x), 'wild_animal'] = 1
sparse.loc[:, 'wild_animal'] = sparse.loc[:, 'wild_animal'].fillna(0)
sparse.loc[sparse.loc[:, 'archive'].apply(lambda x: 'dead_animal' in x), 'dead_animal'] = 1
sparse.loc[:, 'dead_animal'] = sparse.loc[:, 'dead_animal'].fillna(0)
sparse.loc[sparse.loc[:, 'archive'].apply(lambda x: 'leather_object' in x), 'leather_object'] = 1
sparse.loc[:, 'leather_object'] = sparse.loc[:, 'leather_object'].fillna(0)
sparse.loc[sparse.loc[:, 'archive'].apply(lambda x: 'precious_object' in x), 'precious_object'] = 1
sparse.loc[:, 'precious_object'] = sparse.loc[:, 'precious_object'].fillna(0)
sparse.loc[sparse.loc[:, 'archive'].apply(lambda x: 'wool' in x), 'wool'] = 1
sparse.loc[:, 'wool'] = sparse.loc[:, 'wool'].fillna(0)
sparse.head()
| ki | kišib | udu | itud | mu | ud | ga | sila | šu | mu.DU | maškim | ekišibak | zabardab | u | maš | šag | lugal | mašgal | kir | a | en | ensik | egia | igikar | ŋiri | ragaba | dubsar | mašda | saŋŋa | amar | mada | akiti | lu | ab | gud | ziga | uzud | ašgar | gukkal | šugid | ... | niŋna | sikiduʾa | gudumdum | šuhugari | šutur | gaguru | nindašura | ekaskalak | usaŋ | nammah | egizid | nisku | gara | saŋ.DUN₃ | muhaldimgal | šagiagal | šagiamah | kurunakgal | ugulaʾek | šidimgal | kalam | enkud | in | kiʾana | bahar | hurizum | lagab | ibadu | balla | šembulug | li | niŋsaha | ensi | archive | domesticated_animal | wild_animal | dead_animal | leather_object | precious_object | wool | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pn | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 100041 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal} | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100189 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {dead_animal} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 100190 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {dead_animal} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 100191 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {dead_animal} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 100211 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {dead_animal} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
5 rows × 1083 columns
known = sparse.loc[sparse['archive'].apply(len) == 1, :]
unknown = sparse.loc[(sparse['archive'].apply(len) == 0) | (sparse['archive'].apply(len) > 1), :]
unknown_0 = sparse.loc[(sparse['archive'].apply(len) == 0), :]
unknown.shape
(3243, 1083)
1.3 Data Exploration¶
unknown.loc[sparse['archive'].apply(len) > 1, :]
| ki | kišib | udu | itud | mu | ud | ga | sila | šu | mu.DU | maškim | ekišibak | zabardab | u | maš | šag | lugal | mašgal | kir | a | en | ensik | egia | igikar | ŋiri | ragaba | dubsar | mašda | saŋŋa | amar | mada | akiti | lu | ab | gud | ziga | uzud | ašgar | gukkal | šugid | ... | niŋna | sikiduʾa | gudumdum | šuhugari | šutur | gaguru | nindašura | ekaskalak | usaŋ | nammah | egizid | nisku | gara | saŋ.DUN₃ | muhaldimgal | šagiagal | šagiamah | kurunakgal | ugulaʾek | šidimgal | kalam | enkud | in | kiʾana | bahar | hurizum | lagab | ibadu | balla | šembulug | li | niŋsaha | ensi | archive | domesticated_animal | wild_animal | dead_animal | leather_object | precious_object | wool | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pn | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 100217 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100229 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100284 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100330 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100749 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 500210 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 507968 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 509325 | 1.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 512780 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {leather_object, wool} | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 |
| 512812 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
1195 rows × 1083 columns
#find rows where archive has empty set
unknown[unknown['archive'] == set()]
| ki | kišib | udu | itud | mu | ud | ga | sila | šu | mu.DU | maškim | ekišibak | zabardab | u | maš | šag | lugal | mašgal | kir | a | en | ensik | egia | igikar | ŋiri | ragaba | dubsar | mašda | saŋŋa | amar | mada | akiti | lu | ab | gud | ziga | uzud | ašgar | gukkal | šugid | ... | niŋna | sikiduʾa | gudumdum | šuhugari | šutur | gaguru | nindašura | ekaskalak | usaŋ | nammah | egizid | nisku | gara | saŋ.DUN₃ | muhaldimgal | šagiagal | šagiamah | kurunakgal | ugulaʾek | šidimgal | kalam | enkud | in | kiʾana | bahar | hurizum | lagab | ibadu | balla | šembulug | li | niŋsaha | ensi | archive | domesticated_animal | wild_animal | dead_animal | leather_object | precious_object | wool | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pn | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 100292 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100301 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100375 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100376 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 100377 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 519647 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 519650 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 519658 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 519957 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 519959 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
2048 rows × 1083 columns
known_copy = known
known_archives = [known_copy['archive'].to_list()[i].pop() for i in range(len(known_copy['archive'].to_list()))]
known_archives
['domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'precious_object',
'precious_object',
'precious_object',
'precious_object',
'wool',
'wool',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'leather_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'wool',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'leather_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'wild_animal',
'wild_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'wool',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'precious_object',
'domesticated_animal',
'precious_object',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'precious_object',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'precious_object',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'precious_object',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'wild_animal',
'precious_object',
'dead_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'precious_object',
'domesticated_animal',
'dead_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'domesticated_animal',
'dead_animal',
'wild_animal',
'domesticated_animal',
'domesticated_animal',
...]
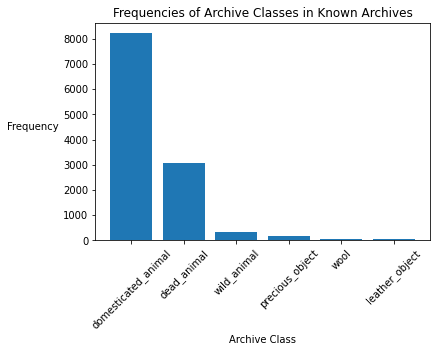
known['archive_class'] = known_archives
archive_counts = known['archive_class'].value_counts()
plt.xlabel('Archive Class')
plt.ylabel('Frequency', rotation=0, labelpad=30)
plt.title('Frequencies of Archive Classes in Known Archives')
plt.xticks(rotation=45)
plt.bar(archive_counts.index, archive_counts);
percent_domesticated_animal = archive_counts['domesticated_animal'] / sum(archive_counts)
print('Percent of texts in Domesticated Animal Archive:', percent_domesticated_animal)
Percent of texts in Domesticated Animal Archive: 0.6905682582380632
known.shape
(11896, 1084)
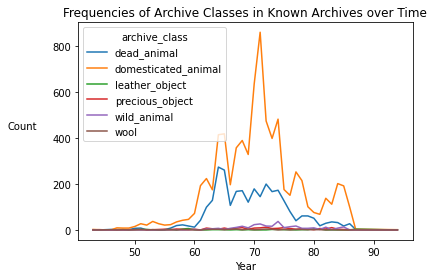
words_df_copy = words_df.copy()
words_df_copy['id_text'] = [int(pn[1:]) for pn in words_df_copy['id_text']]
grouped = words_df_copy.groupby(by=['id_text']).first()
grouped = grouped.fillna(0)
known_copy = known.copy()
known_copy['year'] = grouped.loc[grouped.index.isin(known.index),:]['min_year']
year_counts = known_copy.groupby(by=['year', 'archive_class'], as_index=False).count().set_index('year').loc[:, 'archive_class':'ki']
year_counts_pivoted = year_counts.pivot(columns='archive_class', values='ki').fillna(0)
year_counts_pivoted.drop(index=0).plot();
plt.xlabel('Year')
plt.ylabel('Count', rotation=0, labelpad=30)
plt.title('Frequencies of Archive Classes in Known Archives over Time');
known
| ki | kišib | udu | itud | mu | ud | ga | sila | šu | mu.DU | maškim | ekišibak | zabardab | u | maš | šag | lugal | mašgal | kir | a | en | ensik | egia | igikar | ŋiri | ragaba | dubsar | mašda | saŋŋa | amar | mada | akiti | lu | ab | gud | ziga | uzud | ašgar | gukkal | šugid | ... | sikiduʾa | gudumdum | šuhugari | šutur | gaguru | nindašura | ekaskalak | usaŋ | nammah | egizid | nisku | gara | saŋ.DUN₃ | muhaldimgal | šagiagal | šagiamah | kurunakgal | ugulaʾek | šidimgal | kalam | enkud | in | kiʾana | bahar | hurizum | lagab | ibadu | balla | šembulug | li | niŋsaha | ensi | archive | domesticated_animal | wild_animal | dead_animal | leather_object | precious_object | wool | archive_class | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pn | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 100041 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 100189 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | dead_animal |
| 100190 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | dead_animal |
| 100191 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | dead_animal |
| 100211 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | dead_animal |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 514376 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 517184 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | {} | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | dead_animal |
| 519457 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 519521 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | wild_animal |
| 519792 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
11896 rows × 1084 columns
#run to save the prepared data
known.to_csv('output/part_4_known.csv')
known.to_pickle('output/part_4_known.p')
unknown.to_csv('output/part_4_unknown.csv')
unknown.to_pickle('output/part_4_unknown.p')
unknown_0.to_csv('output/part_4_unknown_0.csv')
unknown_0.to_pickle('output/part_4_unknown_0.p')
#known = pd.read_pickle('https://gitlab.com/yashila.bordag/sumnet-data/-/raw/main/part_4_known.p')
#unknown = pd.read_pickle('https://gitlab.com/yashila.bordag/sumnet-data/-/raw/main/part_4_unknown.p')
#unknown_0 = pd.read_pickle('https://gitlab.com/yashila.bordag/sumnet-data/-/raw/main/part_4_unknown_0.p')
model_weights = {}
1.3.1 PCA/Dimensionality Reduction¶
Here we perform PCA to find out more about the underlying structure of the dataset. We will analyze the 2 most important principle components and explore how much of the variation of the known set is due to these components.
#PCA
pca_archive = PCA()
principalComponents_archive = pca_archive.fit_transform(known.loc[:, 'AN.bu.um':'šuʾura'])
principal_archive_Df = pd.DataFrame(data = principalComponents_archive
, columns = ['principal component ' + str(i) for i in range(1, 1 + len(principalComponents_archive[0]))])
len(known.loc[:, 'AN.bu.um':'šuʾura'].columns)
69
principal_archive_Df
| principal component 1 | principal component 2 | principal component 3 | principal component 4 | principal component 5 | principal component 6 | principal component 7 | principal component 8 | principal component 9 | principal component 10 | principal component 11 | principal component 12 | principal component 13 | principal component 14 | principal component 15 | principal component 16 | principal component 17 | principal component 18 | principal component 19 | principal component 20 | principal component 21 | principal component 22 | principal component 23 | principal component 24 | principal component 25 | principal component 26 | principal component 27 | principal component 28 | principal component 29 | principal component 30 | principal component 31 | principal component 32 | principal component 33 | principal component 34 | principal component 35 | principal component 36 | principal component 37 | principal component 38 | principal component 39 | principal component 40 | principal component 41 | principal component 42 | principal component 43 | principal component 44 | principal component 45 | principal component 46 | principal component 47 | principal component 48 | principal component 49 | principal component 50 | principal component 51 | principal component 52 | principal component 53 | principal component 54 | principal component 55 | principal component 56 | principal component 57 | principal component 58 | principal component 59 | principal component 60 | principal component 61 | principal component 62 | principal component 63 | principal component 64 | principal component 65 | principal component 66 | principal component 67 | principal component 68 | principal component 69 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | -5.157586e-17 | 2.706112e-15 | -0.000481 | -0.000306 | 0.000033 | -3.429768e-16 | 5.923137e-17 | 4.357782e-17 | -2.465750e-16 | 2.860153e-16 | -0.000624 | 3.966646e-17 | 2.603275e-16 | -4.583354e-17 | -2.646101e-17 | 8.451599e-18 | -3.045379e-17 | 1.346336e-17 | 9.688025e-17 | 1.936758e-17 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -1.617357e-16 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | 1.970750e-15 | 1.869525e-17 | 9.415297e-16 | 2.019903e-15 | -1.169278e-18 | 1.894373e-20 | -7.211354e-21 | -2.128106e-20 | 1.746307e-21 | -2.687171e-20 | -2.849700e-20 | 2.445157e-21 | 1.963535e-20 | 3.486655e-21 | -2.377182e-22 | 1.506376e-20 | 1.319668e-21 | -2.913316e-23 | 4.035962e-23 | -2.844907e-23 | 5.776068e-23 | -3.962093e-21 |
| 1 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | 1.564922e-16 | 2.649608e-15 | -0.000481 | -0.000306 | 0.000033 | -2.727474e-16 | -3.278168e-17 | -2.041174e-17 | 1.211487e-16 | 4.041074e-17 | -0.000624 | 1.172185e-16 | 2.181246e-17 | 3.816922e-17 | -1.415496e-17 | -8.185121e-17 | 1.835050e-16 | -2.723499e-16 | 1.982747e-16 | -1.303244e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -6.575466e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | 9.619220e-15 | -6.600049e-17 | -1.429446e-16 | -1.916333e-16 | -4.540458e-17 | 3.457458e-19 | -6.477865e-19 | 3.464390e-19 | -1.141279e-19 | 2.092593e-19 | 1.045044e-18 | 1.123598e-18 | -1.462768e-18 | 1.294085e-18 | 4.663841e-19 | -3.597813e-19 | -4.046188e-20 | -1.200163e-20 | -1.178408e-20 | -6.275206e-21 | -1.761853e-20 | 6.692983e-20 |
| 2 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | 1.535254e-16 | 2.658846e-15 | -0.000481 | -0.000306 | 0.000033 | -2.825484e-16 | -3.280272e-17 | -2.539356e-17 | 1.231791e-16 | 4.753436e-17 | -0.000624 | 1.205493e-16 | 2.333757e-17 | 3.548553e-17 | -1.480301e-17 | -8.340641e-17 | 1.838694e-16 | -2.681072e-16 | 1.870916e-16 | -1.443350e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -6.308277e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | 9.504357e-15 | -6.459051e-17 | -1.301681e-16 | -1.933703e-16 | -3.696746e-17 | 1.681898e-19 | -6.751741e-19 | 4.477259e-19 | -7.269816e-20 | 4.045474e-19 | 1.223637e-18 | 1.075851e-18 | -1.593793e-18 | 1.216670e-18 | 2.723570e-19 | -4.703521e-19 | 3.232220e-19 | 7.222399e-21 | 5.820028e-21 | 5.370710e-21 | 2.005591e-20 | 1.115431e-19 |
| 3 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | -9.229732e-18 | 2.727819e-15 | -0.000481 | -0.000306 | 0.000033 | -2.245254e-16 | -2.029954e-17 | -1.117705e-17 | -8.220052e-17 | 5.707972e-17 | -0.000624 | -3.630915e-18 | 1.008721e-16 | 8.512500e-19 | 4.287588e-18 | -4.577265e-18 | -8.689851e-19 | -1.644222e-17 | 4.959985e-17 | -1.224857e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -2.502096e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | -3.722096e-18 | -1.397288e-18 | 2.365451e-19 | 5.917212e-21 | -1.645157e-19 | -4.078526e-21 | -4.933178e-20 | 2.974763e-20 | -5.485142e-20 | -1.298604e-19 | -6.587155e-20 | -1.116076e-19 | 1.338942e-19 | 8.165997e-20 | 1.384465e-19 | -1.084071e-20 | 9.970619e-21 | -4.728920e-20 | -4.616010e-20 | -2.324160e-20 | -6.416898e-20 | -3.414129e-20 |
| 4 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | 1.769313e-18 | 2.735146e-15 | -0.000481 | -0.000306 | 0.000033 | -1.948217e-16 | -3.447135e-17 | -4.074711e-17 | -7.168826e-17 | 4.512882e-17 | -0.000624 | 1.529291e-17 | 1.304296e-16 | -1.218269e-18 | 6.169002e-18 | -1.717860e-17 | -2.588438e-17 | -2.077230e-17 | 3.981505e-17 | -1.113311e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -2.206909e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | -2.521881e-17 | -1.442576e-17 | -1.046305e-16 | 3.904241e-17 | 1.555701e-17 | 1.626597e-17 | 2.837998e-17 | 1.654359e-18 | -5.697258e-19 | 1.987760e-17 | 8.221763e-18 | 1.752222e-17 | -1.942312e-17 | -1.099805e-18 | 3.031280e-17 | -4.952675e-21 | -1.094244e-19 | -2.285040e-20 | -3.140286e-20 | -8.951914e-21 | -3.201393e-20 | 9.188216e-19 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 11879 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | -4.823243e-18 | 2.730673e-15 | -0.000481 | -0.000306 | 0.000033 | -2.266430e-16 | -1.855078e-17 | -1.093610e-17 | -7.766431e-17 | 5.736404e-17 | -0.000624 | -1.946653e-18 | 1.082104e-16 | -9.088522e-18 | 5.251149e-19 | -3.071913e-18 | 2.347497e-18 | -5.358961e-18 | 5.214637e-17 | -1.227922e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -2.485590e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | -1.981995e-20 | -1.408317e-20 | 4.777440e-21 | -1.768909e-21 | -3.751041e-21 | 1.166248e-20 | -8.218202e-21 | -2.474420e-20 | 2.830425e-20 | 4.740412e-20 | -3.458524e-20 | 7.981751e-21 | -3.462157e-20 | -1.215977e-20 | -2.985825e-20 | 6.461742e-21 | -1.206271e-20 | 8.583833e-22 | 1.080165e-21 | 9.273926e-23 | -1.733363e-20 | 3.412558e-22 |
| 11880 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | -4.749602e-18 | 2.730674e-15 | -0.000481 | -0.000306 | 0.000033 | -2.266228e-16 | -1.869866e-17 | -1.114803e-17 | -7.765207e-17 | 5.744572e-17 | -0.000624 | -1.983822e-18 | 1.082685e-16 | -8.996091e-18 | 5.472673e-19 | -3.003457e-18 | 2.314899e-18 | -5.345549e-18 | 5.225182e-17 | -1.227673e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -2.485989e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | -1.651102e-20 | -8.997080e-21 | -1.231869e-19 | 4.509590e-20 | 1.841453e-20 | -1.625999e-20 | 2.114757e-20 | 4.172252e-20 | -7.715687e-20 | -2.953932e-20 | 3.815738e-20 | -7.018896e-20 | 5.159629e-20 | 6.339990e-20 | 9.522999e-20 | -2.781818e-20 | 3.738191e-19 | -8.328490e-21 | -9.461461e-21 | -1.915426e-21 | -1.542882e-20 | -1.716732e-21 |
| 11881 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | -4.745834e-18 | 2.730674e-15 | -0.000481 | -0.000306 | 0.000033 | -2.266228e-16 | -1.869866e-17 | -1.114775e-17 | -7.765208e-17 | 5.744572e-17 | -0.000624 | -1.984056e-18 | 1.082685e-16 | -8.996113e-18 | 5.469898e-19 | -3.003380e-18 | 2.314654e-18 | -5.345462e-18 | 5.225182e-17 | -1.227673e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -2.485053e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | -1.648348e-20 | -8.950326e-21 | -1.263781e-19 | 4.626611e-20 | 1.896135e-20 | -1.660133e-20 | 2.106983e-20 | 4.160353e-20 | -7.701988e-20 | -2.993905e-20 | 3.810188e-20 | -7.045446e-20 | 5.129683e-20 | 6.345979e-20 | 9.529311e-20 | -2.772713e-20 | 3.735923e-19 | -9.942613e-21 | -1.135000e-20 | -2.100779e-21 | 1.085541e-20 | -6.187519e-22 |
| 11882 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | -4.745834e-18 | 2.730674e-15 | -0.000481 | -0.000306 | 0.000033 | -2.266228e-16 | -1.869866e-17 | -1.114775e-17 | -7.765208e-17 | 5.744572e-17 | -0.000624 | -1.984056e-18 | 1.082685e-16 | -8.996113e-18 | 5.469898e-19 | -3.003380e-18 | 2.314654e-18 | -5.345462e-18 | 5.225182e-17 | -1.227673e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -2.485053e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | -1.648348e-20 | -8.950326e-21 | -1.263781e-19 | 4.626611e-20 | 1.896135e-20 | -1.660133e-20 | 2.106983e-20 | 4.160353e-20 | -7.701988e-20 | -2.993905e-20 | 3.810188e-20 | -7.045446e-20 | 5.129683e-20 | 6.345979e-20 | 9.529311e-20 | -2.772713e-20 | 3.735923e-19 | -9.942613e-21 | -1.135000e-20 | -2.100779e-21 | 1.085541e-20 | -6.187519e-22 |
| 11883 | -0.001153 | -0.000651 | -0.000559 | -0.000524 | 0.00003 | 0.000357 | -0.000339 | -0.000428 | -0.000424 | -0.000372 | -0.000382 | -4.745834e-18 | 2.730674e-15 | -0.000481 | -0.000306 | 0.000033 | -2.266228e-16 | -1.869866e-17 | -1.114775e-17 | -7.765208e-17 | 5.744572e-17 | -0.000624 | -1.984056e-18 | 1.082685e-16 | -8.996113e-18 | 5.469898e-19 | -3.003380e-18 | 2.314654e-18 | -5.345462e-18 | 5.225182e-17 | -1.227673e-16 | -0.00051 | 0.000105 | -0.000049 | -0.000055 | -0.000027 | -0.000113 | -0.000011 | -0.000028 | -0.000117 | -2.485053e-17 | -0.000085 | -0.000069 | -0.000107 | 0.000015 | -0.000031 | -0.000042 | -1.648348e-20 | -8.950326e-21 | -1.263781e-19 | 4.626611e-20 | 1.896135e-20 | -1.660133e-20 | 2.106983e-20 | 4.160353e-20 | -7.701988e-20 | -2.993905e-20 | 3.810188e-20 | -7.045446e-20 | 5.129683e-20 | 6.345979e-20 | 9.529311e-20 | -2.772713e-20 | 3.735923e-19 | -9.942613e-21 | -1.135000e-20 | -2.100779e-21 | 1.085541e-20 | -6.187519e-22 |
11884 rows × 69 columns
principal_archive_Df.shape
(11884, 69)
print('Explained variation per principal component: {}'.format(pca_archive.explained_variance_ratio_))
Explained variation per principal component: [1.48909600e-01 7.68896157e-02 5.44580086e-02 3.84859466e-02
3.73441272e-02 3.55736929e-02 3.34908306e-02 3.23180930e-02
3.08607958e-02 2.72465476e-02 2.62104855e-02 2.46991069e-02
2.46991069e-02 2.46822947e-02 2.23337377e-02 1.90080231e-02
1.85243302e-02 1.85243302e-02 1.85243302e-02 1.85243302e-02
1.85243302e-02 1.84959965e-02 1.23495535e-02 1.23495535e-02
1.23495535e-02 1.23495535e-02 1.23495535e-02 1.23495535e-02
1.23495535e-02 1.23495535e-02 1.23495535e-02 1.23306530e-02
1.17690579e-02 1.08917149e-02 1.05830913e-02 9.79182442e-03
9.66057258e-03 8.53332346e-03 8.13466952e-03 7.97584503e-03
6.17477673e-03 6.17425099e-03 4.86154090e-03 4.81603581e-03
4.71709331e-03 2.85814508e-03 1.25376384e-03 1.15905668e-30
3.84394984e-31 3.83344024e-32 3.01169726e-32 4.83032948e-33
7.20704280e-34 7.20704280e-34 7.20704280e-34 7.20704280e-34
7.20704280e-34 7.20704280e-34 7.20704280e-34 7.20704280e-34
7.20704280e-34 7.20704280e-34 7.20704280e-34 7.20704280e-34
7.20704280e-34 7.20704280e-34 7.20704280e-34 7.20704280e-34
1.58880404e-34]
plt.plot(pca_archive.explained_variance_ratio_)
[<matplotlib.lines.Line2D at 0x7f075bc2b510>]
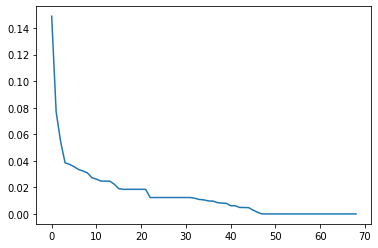
known_reindexed = known.reset_index()
known_reindexed
plt.figure()
plt.figure(figsize=(10,10))
plt.xticks(fontsize=12)
plt.yticks(fontsize=14)
plt.xlabel('Principal Component 1',fontsize=20)
plt.ylabel('Principal Component 2',fontsize=20)
plt.title("Principal Component Analysis of Archives",fontsize=20)
targets = ['domesticated_animal', 'wild_animal', 'dead_animal', 'leather_object', 'precious_object', 'wool']
colors = ['red', 'orange', 'yellow', 'green', 'blue', 'violet']
for target, color in zip(targets,colors):
indicesToKeep = known_reindexed.index[known_reindexed['archive_class'] == target].tolist()
plt.scatter(principal_archive_Df.loc[indicesToKeep, 'principal component 1']
, principal_archive_Df.loc[indicesToKeep, 'principal component 2'], c = color, s = 50)
plt.legend(targets,prop={'size': 15})
# import seaborn as sns
# plt.figure(figsize=(16,10))
# sns.scatterplot(
# x="principal component 1", y="principal component 2",
# hue="y",
# palette=sns.color_palette("hls", 10),
# data=principal_cifar_Df,
# legend="full",
# alpha=0.3
# )
2 Simple Modeling Methods¶
2.1 Logistic Regression¶
Here we will train our model using logistic regression to predict archives based on the features made in the previous subsection.
2.1.1 Logistic Regression by Archive¶
Here we will train and test a set of 1 vs all Logistic Regression Classifiers which will attempt to classify tablets as either a part of an archive, or not in an archive.
clf_da = LogisticRegression(random_state=42, solver='lbfgs', max_iter=200)
clf_da.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'domesticated_animal'])
clf_da.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'domesticated_animal'])
clf_wa = LogisticRegression(random_state=42, solver='lbfgs', max_iter=200)
clf_wa.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'wild_animal'])
clf_wa.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'wild_animal'])
clf_dea = LogisticRegression(random_state=42, solver='lbfgs', max_iter=200)
clf_dea.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'dead_animal'])
clf_dea.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'dead_animal'])
clf_lo = LogisticRegression(random_state=42, solver='lbfgs', max_iter=200)
clf_lo.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'leather_object'])
clf_lo.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'leather_object'])
clf_po = LogisticRegression(random_state=42, solver='lbfgs', max_iter=200)
clf_po.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'precious_object'])
clf_po.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'precious_object'])
clf_w = LogisticRegression(random_state=42, solver='lbfgs', max_iter=200)
clf_w.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'wool'])
clf_w.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'wool'])
known.loc[:, 'AN.bu.um':'šuʾura']
As we can see the domesticated animal model has the lowest accuracy while the leather object, precious_object, and wool classifiers work fairly well.
2.1.2 Multinomial Logistic Regression¶
Here we will be using multinomial logistic regression as we have multiple archive which we could classify each text into. We are fitting our data onto the tablets with known archives and then checking the score to see how accurate the model is.
Finally, we append the Logistic Regression prediction as an archive prediction for the tablets without known archives.
clf_archive = LogisticRegression(random_state=42, solver='lbfgs', max_iter=300)
clf_archive.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'archive_class'])
log_reg_score = clf_archive.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'archive_class'])
model_weights['LogReg'] = log_reg_score
log_reg_score
0.6918291862811029
#Predictions for Unknown
unknown["LogReg Predicted Archive"] = clf_archive.predict(unknown.loc[:, 'AN.bu.um':'šuʾura'])
unknown
| ki | kišib | udu | itud | mu | ud | ga | sila | šu | mu.DU | maškim | ekišibak | zabardab | u | maš | šag | lugal | mašgal | kir | a | en | ensik | egia | igikar | ŋiri | ragaba | dubsar | mašda | saŋŋa | amar | mada | akiti | lu | ab | gud | ziga | uzud | ašgar | gukkal | šugid | ... | sikiduʾa | gudumdum | šuhugari | šutur | gaguru | nindašura | ekaskalak | usaŋ | nammah | egizid | nisku | gara | saŋ.DUN₃ | muhaldimgal | šagiagal | šagiamah | kurunakgal | ugulaʾek | šidimgal | kalam | enkud | in | kiʾana | bahar | hurizum | lagab | ibadu | balla | šembulug | li | niŋsaha | ensi | archive | domesticated_animal | wild_animal | dead_animal | leather_object | precious_object | wool | LogReg Predicted Archive | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pn | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 100217 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 100229 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 100284 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {domesticated_animal, wild_animal} | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 100292 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 100301 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 519647 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 519650 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 519658 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 519957 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
| 519959 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | {} | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | domesticated_animal |
3243 rows × 1084 columns
known['archive_class'].unique()
2.2 K Nearest Neighbors¶
Here we will train our model using k nearest neighbors to predict archives based on the features made in the previous subsection. We are fitting our data onto the tablets with known archives and then checking the score to see how accurate the model is.
We then append the KNN prediction as an archive prediction for the tablets without known archives.
Then, we use different values for K (the number of neighbors we take into consideration when predicting for a tablet) to see how the accuracy changes for different values of K. This can be seen as a form of hyperparameter tuning because we are trying to see which K we should choose to get the highest training accuracy.
#takes long time to run, so don't run again
list_k = [3, 5, 7, 9, 11, 13]
max_k, max_score = 0, 0
for k in list_k:
knn = KNeighborsClassifier(n_neighbors=k)
knn.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'archive_class'])
knn_score = knn.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'archive_class'])
print("Accuracy for k = %s: " %(k), knn_score)
if max_score <= knn_score:
max_score = knn_score
max_k = k
As we can see here, k = 5 and k = 9 have the best training accuracy performance which falls roughly in line with the Logistic Regression classification training accuracy.
knn = KNeighborsClassifier(n_neighbors=max_k)
knn.fit(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'archive_class'])
knn_score = knn.score(known.loc[:, 'AN.bu.um':'šuʾura'], known.loc[:, 'archive_class'])
model_weights['KNN'] = knn_score
#Predictions for Unknown
unknown["KNN Predicted Archive"] = knn.predict(unknown.loc[:, 'AN.bu.um':'šuʾura'])
unknown
As we can see in the output from the previous cell, we can get different predictions depending on the classifier we choose.
Next we will split the data we have on tablets with known archives into a test and training set to further understant the atraining accuracy. For the next two sections, we will use X_train and y_train to train the data and X_test and y_test to test the data. As the known set was split randomly, we presume that both the training and test set are representative of the whole known set, so the two sets are reasonably comparable.
#Split known into train and test, eventually predict with unknown
X_train, X_test, y_train, y_test = train_test_split(known.loc[:, 'AN.bu.um':'šuʾura'],
known.loc[:, 'archive_class'],
test_size=0.2,random_state=0)
2.3 Naive Bayes¶
Here we will train our model using a Naive Bayes Model to predict archives based on the features made in the previous subsection. Here, we make the assumption that the features are independent of each other, from which the descriptor naive comes from. So:
and:
Moreover, we will be using Bayes’ Law, which in this case states:
eg. the probability of a particular tablet (defined by features \(x_1, x_2, ..., x_n\)) is in archive \(y\), is equal to the probability of getting a tablet from archive \(y\) times the probability you would get a particular set of features \(x_1, x_2, ..., x_n\) divided by the probability of getting a particular set of features \(x_1, x_2, ..., x_n\).
Applying our assumption of independence from before, we can simplify this to:
Which means the probability of a particular tablet (defined by features \(x_1, x_2, ..., x_n\)) is in archive \(y\) is proportional to
We are training two models where the first assumes the features are Gaussian random variables and the second assumes the features are Bernoulli random variables.
We are fitting our data onto the tablets with known archives and then checking the score to see how accurate the model is.
Finally, we append the two Naive Bayes predictions as archive predictions for the tablets without known archives.
#Gaussian
gauss = GaussianNB()
gauss.fit(X_train, y_train)
gauss_nb_score = gauss.score(X_test, y_test)
model_weights['GaussNB'] = gauss_nb_score
gauss_nb_score
We can see than the Gaussian assumption does quite poorly.
#Predictions for Unknown
unknown["GaussNB Predicted Archive"] = gauss.predict(unknown.loc[:, 'AN.bu.um':'šuʾura'])
unknown
#Bernoulli
bern = BernoulliNB()
bern.fit(X_train, y_train)
bern_nb_score = bern.score(X_test, y_test)
model_weights['BernoulliNB'] = bern_nb_score
bern_nb_score
However the Bernoulli assumption does quite well.
#Predictions for Unknown
unknown["BernoulliNB Predicted Archive"] = bern.predict(unknown.loc[:, 'AN.bu.um':'šuʾura'])
unknown
2.4 SVM¶
Here we will train our model using Support Vector Machines to predict archives based on the features made earlier in this section. We are fitting our data onto the tablets with known archives and then checking the score to see how accurate the model is.
Finally, we append the SVM prediction as an archive prediction for the tablets without known archives.
svm_archive = svm.SVC(kernel='linear')
svm_archive.fit(X_train, y_train)
y_pred = svm_archive.predict(X_test)
svm_score = metrics.accuracy_score(y_test, y_pred)
model_weights['SVM'] = svm_score
print("Accuracy:", svm_score)
unknown["SVM Predicted Archive"] = svm_archive.predict(unknown.loc[:, 'AN.bu.um':'šuʾura'])
unknown
3 Complex Modeling Methods¶
4 Voting Mechanism Between Models¶
Here we will use the models to determine the archive which to assign to each tablet with an unknown archive.
We will then augment the words_df with these archives.
model_weights
def visualize_archives(data, prediction_name):
archive_counts = data.value_counts()
plt.xlabel('Archive Class')
plt.ylabel('Frequency', rotation=0, labelpad=30)
plt.title('Frequencies of ' + prediction_name + ' Predicted Archives')
plt.xticks(rotation=45)
plt.bar(archive_counts.index, archive_counts);
percent_domesticated_animal = archive_counts['domesticated_animal'] / sum(archive_counts)
print('Percent of texts in Domesticated Animal Archive:', percent_domesticated_animal)
#Log Reg Predictions
visualize_archives(unknown['LogReg Predicted Archive'], 'Logistic Regression')
#KNN Predictions
visualize_archives(unknown['KNN Predicted Archive'], 'K Nearest Neighbors')
#Gaussian Naive Bayes Predictions
visualize_archives(unknown['GaussNB Predicted Archive'], 'Gaussian Naive Bayes')
#Bernoulli Naive Bayes Predictions
visualize_archives(unknown['BernoulliNB Predicted Archive'], 'Bernoulli Naive Bayes')
#SVM Predictions
visualize_archives(unknown['SVM Predicted Archive'], 'Support Vector Machines Naive Bayes')
def weighted_voting(row):
votes = {} # create empty voting dictionary
# tally votes
for model in row.index:
model_name = model[:-18] # remove ' Predicted Archive' from column name
prediction = row[model]
if prediction not in votes.keys():
votes[prediction] = model_weights[model_name] # if the prediction isn't in the list of voting categories, add it with a weight equal to the current model weight
else:
votes[prediction] += model_weights[model_name] # else, add model weight to the prediction
return max(votes, key=votes.get) # use the values to get the prediction with the greatest weight
predicted_archives = unknown.loc[:, 'LogReg Predicted Archive':
'SVM Predicted Archive'].copy() # get predictions
weighted_prediction = predicted_archives.apply(weighted_voting, axis=1) #apply voting mechanism on each row and return 'winning' prediction
weighted_prediction[weighted_prediction != 'domesticated_animal']
words_df
archive_class = known['archive_class'].copy().append(weighted_prediction)
words_df['archive_class'] = words_df.apply(lambda row: archive_class[int(row['id_text'][1:])], axis=1)
words_df
5 Sophisticated Naive Bayes¶
import warnings
warnings.filterwarnings('ignore')
5.1 Feature and Model Creation¶
There are some nouns that are so closely associated with a specific archive that their presence in a text virtually guarantees that the text belongs to that archive. We will use this fact to create a training set for our classification model.
The labels dictionary below contains the different archives along with their possible associated nouns.
labels = dict()
labels['domesticated_animal'] = ['ox', 'cow', 'sheep', 'goat', 'lamb', '~sheep', 'equid']
dom = '(' + '|'.join(labels['domesticated_animal']) + ')'
#split domesticated into large and small - sheep, goat, lamb, ~sheep would be small domesticated animals
labels['wild_animal'] = ['bear', 'gazelle', 'mountain', 'lion'] # account for 'mountain animal' and plural
wild = '(' + '|'.join(labels['wild_animal']) + ')'
labels['dead_animal'] = ['die'] # find 'die' before finding domesticated or wild
dead = '(' + '|'.join(labels['dead_animal']) + ')'
labels['leather_object'] = ['boots', 'sandals']
leath = '(' + '|'.join(labels['leather_object']) + ')'
labels['precious_object'] = ['copper', 'bronze', 'silver', 'gold']
prec = '(' + '|'.join(labels['precious_object']) + ')'
labels['wool'] = ['wool', '~wool', 'hair']
wool = '(' + '|'.join(labels['wool']) + ')'
complete = []
for lemma_list in labels.values():
complete = complete + lemma_list
tot = '(' + '|'.join(complete) + ')'
# labels['queens_archive'] = []
dom_tabs = set(words_df.loc[words_df['lemma'].str.match('.*\[.*' + dom + '.*\]')]['id_text'])
wild_tabs = set(words_df.loc[words_df['lemma'].str.match('.*\[.*' + wild + '.*\]')]['id_text'])
dead_tabs = set(words_df.loc[words_df['lemma'].str.match('.*\[.*' + dead + '.*\]')]['id_text'])
leath_tabs = set(words_df.loc[words_df['lemma'].str.match('.*\[.*' + leath + '.*\]')]['id_text'])
prec_tabs = set(words_df.loc[words_df['lemma'].str.match('.*\[.*' + prec + '.*\]')]['id_text'])
wool_tabs = set(words_df.loc[words_df['lemma'].str.match('.*\[.*' + wool + '.*\]')]['id_text'])
Each row of the sparse table below corresponds to one text, and the columns of the table correspond to the words that appear in the texts. Every cell contains the number of times a specific word appears in a certain text.
# remove lemmas that are a part of a seal as well as words that are being used to determine training classes
filter = (~words_df['label'].str.contains('s')) | words_df['lemma'].str.match('.*\[.*' + tot + '.*\]')
sparse = words_df[filter].groupby(by=['id_text', 'lemma']).count()
sparse = sparse['id_word'].unstack('lemma')
sparse = sparse.fillna(0)
#cleaning
del filter
text_length = sparse.sum(axis=1)
If a text contains a word that is one of the designated nouns in labels, it is added to the set to be used for our ML model. Texts that do not contain any of these words or that contain words corresponding to more than one archive are ignored.
class_array = []
for id_text in sparse.index:
cat = None
number = 0
if id_text in dom_tabs:
number += 1
cat = 'dom'
if id_text in wild_tabs:
number += 1
cat = 'wild'
if id_text in dead_tabs:
number += 1
cat = 'dead'
if id_text in prec_tabs:
number += 1
cat = 'prec'
if id_text in wool_tabs:
number += 1
cat = 'wool'
if number == 1:
class_array.append(cat)
else:
class_array.append(None)
class_series = pd.Series(class_array, sparse.index)
Next we remove the texts from sparse that we used in the previous cell.
used_cols = []
for col in sparse.columns:
if re.match('.*\[.*' + tot + '.*\]', col):
used_cols.append(col)
#elif re.match('.*PN$', col) is None:
# used_cols.append(col)
sparse = sparse.drop(used_cols, axis=1)
Now the sparse table will be updated to contain percentages of the frequency that a word appears in the text rather than the raw number of occurrences. This will allow us to better compare frequencies across texts of different lengths.
for col in sparse.columns:
if col != 'text_length':
sparse[col] = sparse[col]/text_length*1000
We must convert percentages from the previous cell into integers for the ML model to work properly.
this sparse = sparse.round()
sparse = sparse.astype(int)
To form X, we reduce the sparse table to only contain texts that were designated for use above in class_series. Y consists of the names of the different archives.
X = sparse.loc[class_series.dropna().index]
X = X.drop(X.loc[X.sum(axis=1) == 0, :].index, axis=0)
y = class_series[X.index]
Our data is split into a training set and a test set. The ML model first uses the training set to learn how to predict the archives for the texts. Afterwards, the test set is used to verify how well our ML model works.
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.4,
random_state = 9)
pipe = Pipeline([
('feature_reduction', SelectPercentile(score_func = f_classif)),
('weighted_multi_nb', MultinomialNB())
])
from sklearn.model_selection import GridSearchCV
f = GridSearchCV(pipe, {
'feature_reduction__percentile' : [i*10 for i in range(1, 10)],
'weighted_multi_nb__alpha' : [i/10 for i in range(1, 10)]
}, verbose = 0, n_jobs = -1)
f.fit(X_train, y_train);
f.best_params_
{'feature_reduction__percentile': 70, 'weighted_multi_nb__alpha': 0.1}
Our best score when run on the training set is about 93.6% accuracy.
f.best_score_
0.9359404096834265
Our best score when run on the test set is very similar to above at 93.2% accuracy, which is good because it suggests that our model isn’t overfitted to only work on the training set.
f.score(X_test, y_test)
0.9321229050279329
predicted = f.predict(sparse)
The predicted_df table is the same as the sparse table from above, except that we have added an extra column at the end named prediction. prediction contains our ML model’s classification of which archive the text belongs to based on the frequency of the words that appear.
predicted_df = sparse.copy()
predicted_df['prediction'] = predicted
predicted_df
| lemma | $AN[NA]NA | $GIR[NA]NA | $HI[NA]NA | $KA[NA]NA | $KI[NA]NA | $LAM[NA]NA | $NI[NA]NA | $UD[NA]NA | $ŠA[NA]NA | $ŠID[NA]NA | 1(aš)-a[]NU | 1(aš)-kam[]NU | 1(aš)-še₃[]NU | 1(aš)[]NU | 1(aš@c)[]NU | 1(ban₂)-bi[]NU | 1(ban₂)-ta[]NU | 1(ban₂)-še₃[]NU | 1(ban₂)[]NU | 1(barig)-ta[]NU | 1(barig)[]NU | 1(burʾu)[]NU | 1(bur₃)[]NU | 1(diš)-a-kam[]NU | 1(diš)-a-še₃[]NU | 1(diš)-a[]NU | 1(diš)-am₃[]NU | 1(diš)-bi[]NU | 1(diš)-kam-aš[]NU | 1(diš)-kam-ma-aš[]NU | 1(diš)-kam-ma[]NU | 1(diš)-kam[]NU | 1(diš)-ta[]NU | 1(diš)-x[]NU | 1(diš)-še₃[]NU | 1(diš)[]NU | 1(diš){ša}[]NU | 1(diš@t)-kam-aš[]NU | 1(diš@t)-kam-ma-aš[]NU | 1(diš@t)-kam[]NU | ... | šuʾi[barber]N | šuʾura[goose]N | ʾa₃-um[NA]NA | Ṣa.lim.tum[00]PN | Ṣe.AŠ₂.da.gan[00]PN | Ṣe.er.ṣe.ra.num₂[00]PN | Ṣe.la[00]PN | Ṣe.li.uš.da.gan[00]PN | Ṣe.lu.uš.da.gan.PA[00]PN | Ṣe.lu.uš[00]PN | Ṣe.lu.uš₂.da.gan[00]PN | Ṣe.ra.am[00]PN | Ṣe.ra[00]PN | Ṣeherkinum[0]PN | Ṣeṣe[0]PN | Ṣe₂.la.šu[00]PN | Ṣi.li.sud₃.da[00]PN | Ṣilala[0]PN | Ṣillašu[0]PN | ṢilliAdad[0]PN | ṢilliSud[0]PN | ṢilliŠulgi[0]PN | ṢillušDagan[0]PN | Ṣillušdagan[0]PN | ṢillušŠulgi[0]PN | Ṣillušṭab[0]PN | Ṣipram[0]PN | Ṣirula[0]PN | Ṣummidili[0]PN | ṣa-bi₂-im[NA]NA | ṣa-bu-um[NA]NA | ṣi-il-x-{d}iškur[NA]NA | ṣi-ip-ra-am[NA]NA | ṣi-ra-am[NA]NA | Ṭabaʾili[0]PN | Ṭabumšar[0]PN | Ṭabumšarri[0]PN | Ṭabši[0]SN | Ṭahili[0]PN | prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id_text | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| P100041 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dom |
| P100189 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dead |
| P100190 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dom |
| P100191 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 48 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dead |
| P100211 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 30 | 0 | 0 | 0 | 152 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dead |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| P519650 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dom |
| P519658 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | wild |
| P519792 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 155 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dom |
| P519957 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 71 | 0 | 71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dom |
| P519959 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | dead |
15132 rows × 9174 columns
predicted_df.index
Index(['P100041', 'P100189', 'P100190', 'P100191', 'P100211', 'P100214',
'P100215', 'P100217', 'P100218', 'P100219',
...
'P519534', 'P519613', 'P519623', 'P519624', 'P519647', 'P519650',
'P519658', 'P519792', 'P519957', 'P519959'],
dtype='object', name='id_text', length=15132)
5.4 Testing the Model on Hand-Classified Data¶
Here we first use our same ML model from before on Niek’s hand-classified texts from the wool archive. Testing our ML model on these tablets gives us 82.5% accuracy.
wool_hand_tabs = set(pd.read_csv('drive/MyDrive/SumerianNetworks/JupyterBook/Outputs/wool_pid.txt',header=None)[0])
hand_wool_frame = sparse.loc[wool_hand_tabs].loc[class_series.isna() == True]
f.score(X = hand_wool_frame,
y = pd.Series(
index = hand_wool_frame.index,
data = ['wool' for i in range(0, hand_wool_frame.shape[0])] ))
0.8253968253968254
Testing our ML model on 100 random hand-classified tablets selected from among all the texts gives us 87.2% accuracy.
niek_100_random_tabs = pd.read_pickle('/content/drive/MyDrive/niek_cats').dropna()
niek_100_random_tabs = niek_100_random_tabs.set_index('pnum')['category_text']
random_frame = sparse.loc[set(niek_100_random_tabs.index)]
random_frame['result'] = niek_100_random_tabs[random_frame.index]
f.score(X=random_frame.drop(labels='result', axis=1), y = random_frame['result'])
0.872093023255814
A large majority of the tablets are part of the domestic archive and have been classified as such.
random_frame['result'].array
<PandasArray>
['dead', 'dom', 'dead', 'dom', 'dom', 'dom', 'dead', 'dom', 'dead',
'dom', 'dead', 'dead', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom',
'dom', 'dom', 'wild', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom',
'dead', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom', 'dead', 'dom',
'dom', 'dom', 'dom', 'dom', 'dom', 'dead', 'dom', 'dom', 'dom',
'dom', 'dom', 'dom', 'dead', 'dom', 'dom', 'dom', 'dom', 'wild',
'dom', 'dom', 'dom', 'dom', 'dom', 'dom', 'dead', 'wild', 'dom',
'dom', 'dom', 'dom', 'wild', 'dom', 'dead', 'dom', 'dead', 'dead',
'dom', 'dead', 'dead', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom',
'dom', 'dom', 'dom', 'dom', 'dom']
Length: 86, dtype: object
f.predict(random_frame.drop(labels='result', axis=1))
array(['dead', 'dom', 'dead', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom',
'dom', 'dead', 'dead', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom',
'dom', 'dom', 'wild', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom',
'dead', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom', 'dead', 'dom',
'dom', 'dom', 'dom', 'dom', 'dom', 'dead', 'dom', 'dom', 'dom',
'wild', 'dom', 'dom', 'dom', 'wild', 'dom', 'dom', 'dom', 'dom',
'dom', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom', 'dead', 'dom',
'dom', 'dom', 'dom', 'dom', 'dom', 'dead', 'dom', 'dead', 'dom',
'dom', 'dom', 'dead', 'dom', 'dom', 'dom', 'dom', 'dom', 'dom',
'dom', 'dom', 'dom', 'dom', 'dom'], dtype='<U4')
5.2 Frequency Graphs¶
When run on all of the tablets, our ML model classifies a large portion of the texts into the domestic archive, since that is the most common one.
from matplotlib import pyplot as plt
plt.xlabel('Archive Class')
plt.ylabel('Frequency', rotation=0, labelpad=30)
plt.title('Frequencies of Predicted Archive Classes in All Tablets')
plt.xticks(rotation=45)
labels = list(set(predicted_df['prediction']))
counts = [predicted_df.loc[predicted_df['prediction'] == label].shape[0] for label in labels]
plt.bar(labels, counts);
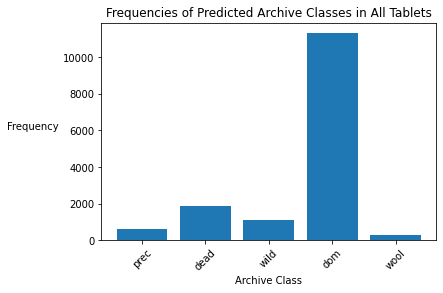
The below chart displays the actual frequencies of the different archives in the test set. As mentioned previously, it is visually obvious that there are many texts in the domestic archive, with comparatively very few texts in all of the other archives.
plt.xlabel('Archive Class')
plt.ylabel('Frequency', rotation=0, labelpad=30)
plt.title('Frequencies of Test Archive Classes')
plt.xticks(rotation=45)
test_counts = [(class_series[X_test.index])[class_series == label].count() for label in labels]
plt.bar(labels, np.asarray(test_counts));
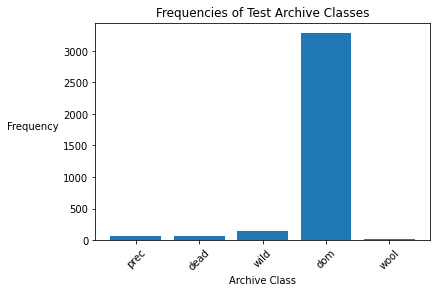
Below is a chart of the predicted frequency of the different archives by our ML model in the test set. Our predicted frequency looks very similar to the actual frequency above, which is good.
plt.xlabel('Archive Class')
plt.ylabel('Frequency', rotation=0, labelpad=30)
plt.title('Frequencies of Predicted Test Archive Classes')
plt.xticks(rotation=45)
test_pred_counts = [predicted_df.loc[X_test.index].loc[predicted_df['prediction'] == label].shape[0] for label in labels]
plt.bar(labels, np.asarray(test_pred_counts));
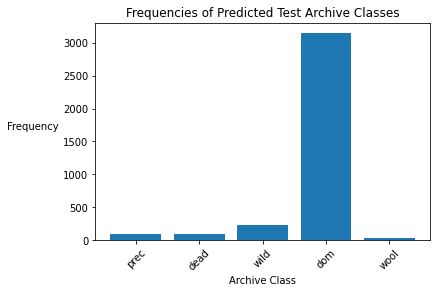
Unfortunately, since our texts skew so heavily towards being part of the domestic archive, most of the other archives end up being overpredicted (i.e. our model says a text is part of that archive when it is actually not). Below we can see that the domestic archive is the only archive whose texts are not overpredicted.
plt.xlabel('Archive Class')
plt.ylabel('Rate', rotation=0, labelpad=30)
plt.title('Rate of Overprediction by Archive')
plt.xticks(rotation=45)
rate = np.asarray(test_pred_counts)/np.asarray(test_counts)*sum(test_counts)/sum(test_pred_counts)
plt.bar(labels, rate);
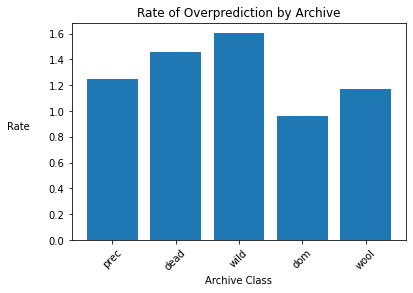
5.3 Accuracy By Archive¶
The accuracies for the dead and wild archives are relatively low. This is likely because those texts are being misclassified into the domestic archive, our largest archive, since all three of these archives deal with animals. The wool and precious archives have decent accuracies.
f.score(X_test[class_series == 'dead'], y_test[class_series == 'dead'])
0.734375
f.score(X_test[class_series == 'dom'], y_test[class_series == 'dom'])
0.9449010654490106
f.score(X_test[class_series == 'wild'], y_test[class_series == 'wild'])
0.7410071942446043
f.score(X_test[class_series == 'wool'], y_test[class_series == 'wool'])
0.8333333333333334
f.score(X_test[class_series == 'prec'], y_test[class_series == 'prec'])
0.9264705882352942
We can also look at the confusion matrix. A confusion matrix is used to evaluate the accuracy of a classification. The rows denote the actual archive, while the columns denote the predicted archive.
Looking at the first column:
73.44% of the dead archive texts are predicted correctly
1.31% of the domestic archive texts are predicted to be part of the dead archive
1.47% of the wild archive texts are predicted to be part of the dead archive
1.43% of the wool archive texts are predicted to be part of the dead archive
none of the precious archive texts are predicted to be part of the dead archive
from sklearn.metrics import confusion_matrix
archive_confusion = confusion_matrix(y_test, f.predict(X_test), normalize='true')
archive_confusion
array([[0.734375 , 0.171875 , 0.015625 , 0.0625 , 0.015625 ],
[0.0130898 , 0.94490107, 0.00487062, 0.03531202, 0.00182648],
[0.01470588, 0.05882353, 0.92647059, 0. , 0. ],
[0.01438849, 0.21582734, 0.02158273, 0.74100719, 0.00719424],
[0. , 0.08333333, 0.08333333, 0. , 0.83333333]])
This is the same confusion matrix converted into real numbers of texts. Since the number of domestic archive texts is so high, even a small bit of misclassification of the domestic archive texts can overwhelm the other archives.
For example, even though only 1.3% of the domestic archive texts are predicted to be part of the dead archive, that corresponds to 43 texts, while the 73% of the dead archive texts that were predicted correctly correspond to just 47 texts. As a result, about half of the texts that were predicted to be part of the dead archive are incorrectly classified.
confusion_matrix(y_test, f.predict(X_test), normalize=None)
array([[ 47, 11, 1, 4, 1],
[ 43, 3104, 16, 116, 6],
[ 1, 4, 63, 0, 0],
[ 2, 30, 3, 103, 1],
[ 0, 2, 2, 0, 20]])